This takes a probsens-family object and produces the distribution plot of
chosen bias parameters, as well as distribution of adjusted measures (with confidence
interval). It can also produce a forest plot of relative risks or odds ratios (with
probsens(), probsens_conf(), or probsens.sel())
Arguments
- x
An object of class "episensr.probsens" returned from the
episensr probsens,probsens.sel,probsens_conf,probsens.irr,probsens.irr.conffunctions.- parms
Choice between adjusted relative risk (
rr) and odds ratio (or), total error relative risk and odds ratio (rr_totandor_tot), forest plots (forest_rrandforest_or),seca,seexp,spca, andspexp,prev.exp,prev.nexpandrisk,irrandirr_tot.- ...
Other unused arguments.
See also
probsens, probsens.sel, probsens_conf,
probsens.irr, probsens.irr.conf
Other visualization:
plot.episensr.booted(),
plot.mbias()
Examples
set.seed(123)
risk <- probsens(matrix(c(45, 94, 257, 945),
dimnames = list(c("BC+", "BC-"), c("Smoke+", "Smoke-")), nrow = 2, byrow = TRUE),
type = "exposure", reps = 20000,
seca = list("trapezoidal", c(.75, .85, .95, 1)),
spca = list("trapezoidal", c(.75, .85, .95, 1)))
#> ℹ Calculating observed measures
#> ⠙ Assign probability distributions
#> ✔ Assign probability distributions [14ms]
#>
#> ⠙ Simple bias analysis
#> ✔ Simple bias analysis [37ms]
#>
#> ⠙ Incorporating random error
#> ! Chosen Se/Sp distributions lead to 821 impossible values which were discarded.
#> ⠙ Incorporating random error
#> ⠹ Incorporating random error
#> ✔ Incorporating random error [138ms]
#>
plot(risk, "rr")
 set.seed(123)
odds <- probsens(matrix(c(45, 94, 257, 945),
dimnames = list(c("BC+", "BC-"), c("Smoke+", "Smoke-")), nrow = 2, byrow = TRUE),
type = "exposure", reps = 20000,
seca = list("beta", c(908, 16)),
seexp = list("beta", c(156, 56)),
spca = list("beta", c(153, 6)),
spexp = list("beta", c(205, 18)),
corr_se = .8,
corr_sp = .8)
#> ℹ Calculating observed measures
#> ⠙ Assign probability distributions
#> ✔ Assign probability distributions [154ms]
#>
#> ⠙ Simple bias analysis
#> ✔ Simple bias analysis [48ms]
#>
#> ⠙ Incorporating random error
#> ⠹ Incorporating random error
#> ✔ Incorporating random error [123ms]
#>
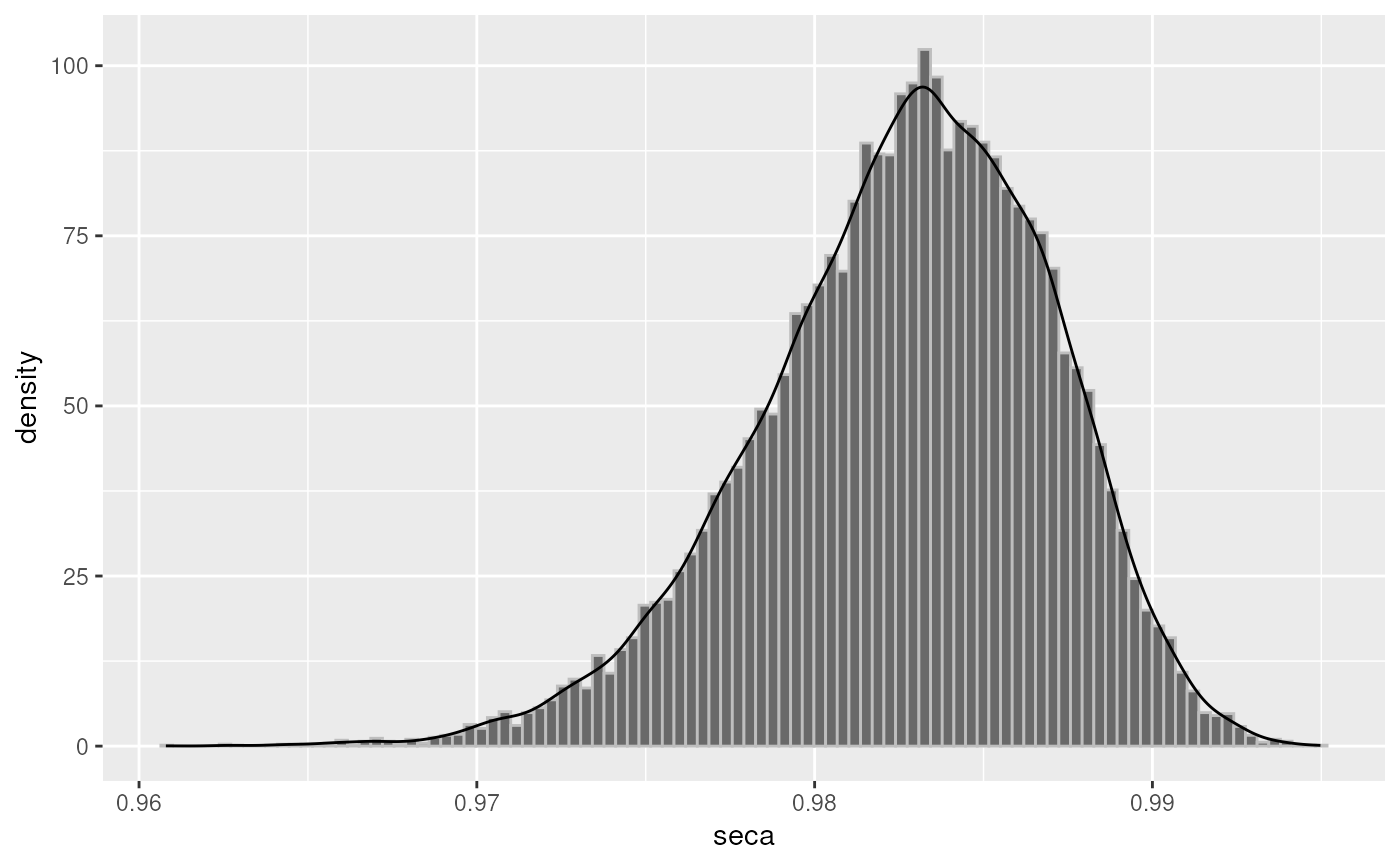
plot(odds, "seca")
set.seed(123)
odds <- probsens(matrix(c(45, 94, 257, 945),
dimnames = list(c("BC+", "BC-"), c("Smoke+", "Smoke-")), nrow = 2, byrow = TRUE),
type = "exposure", reps = 20000,
seca = list("beta", c(908, 16)),
seexp = list("beta", c(156, 56)),
spca = list("beta", c(153, 6)),
spexp = list("beta", c(205, 18)),
corr_se = .8,
corr_sp = .8)
#> ℹ Calculating observed measures
#> ⠙ Assign probability distributions
#> ✔ Assign probability distributions [154ms]
#>
#> ⠙ Simple bias analysis
#> ✔ Simple bias analysis [48ms]
#>
#> ⠙ Incorporating random error
#> ⠹ Incorporating random error
#> ✔ Incorporating random error [123ms]
#>
plot(odds, "seca")
 set.seed(123)
smoke <- probsens(matrix(c(215, 1449, 668, 4296),
dimnames = list(c("BC+", "BC-"), c("Smoke+", "Smoke-")), nrow = 2, byrow = TRUE),
type = "exposure", reps = 20000,
seca = list("uniform", c(.7, .95)),
spca = list("uniform", c(.9, .99)))
#> ℹ Calculating observed measures
#> ⠙ Assign probability distributions
#> ✔ Assign probability distributions [14ms]
#>
#> ⠙ Simple bias analysis
#> ✔ Simple bias analysis [41ms]
#>
#> ⠙ Incorporating random error
#> ⠹ Incorporating random error
#> ✔ Incorporating random error [123ms]
#>
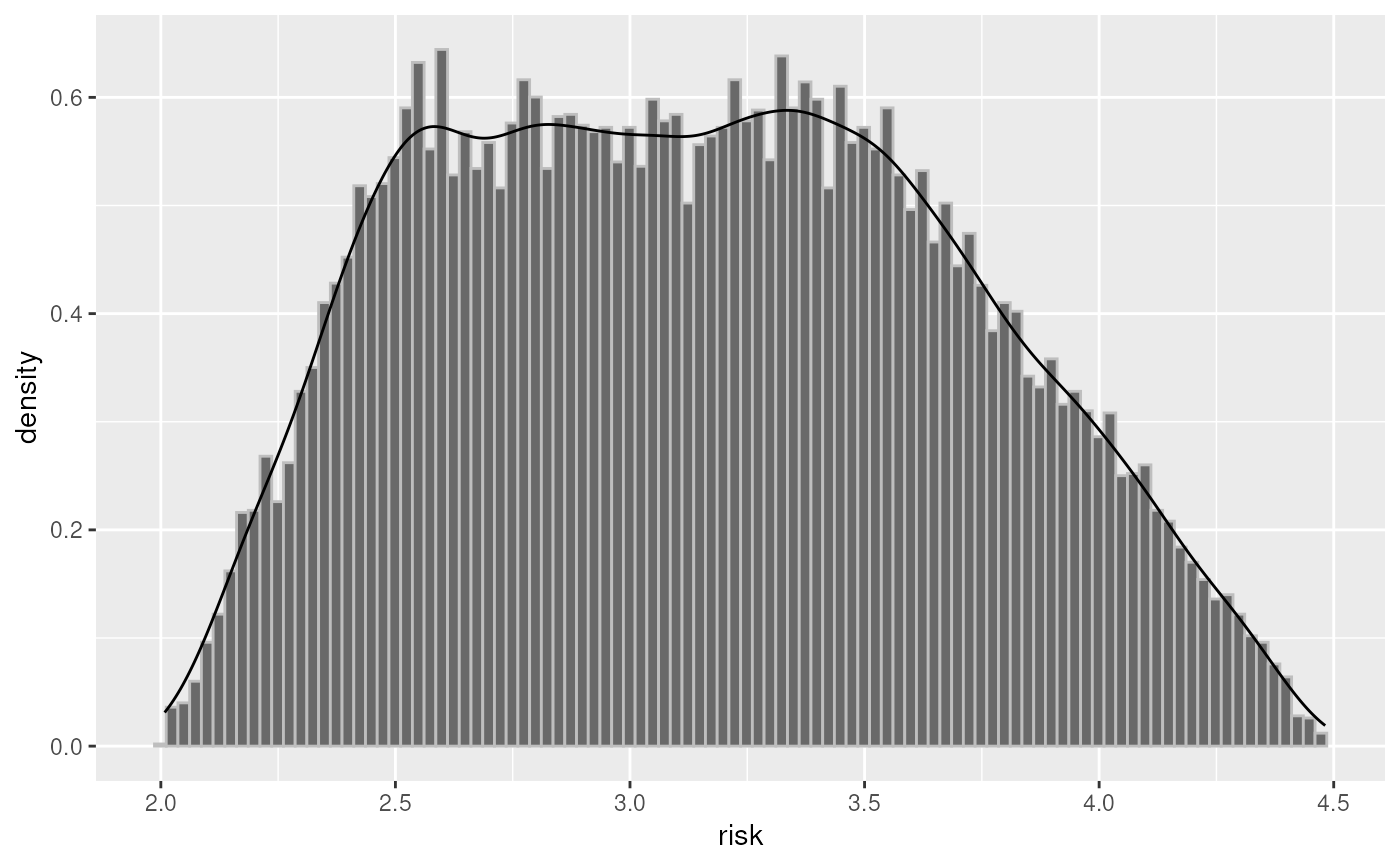
plot(smoke, "forest_or")
set.seed(123)
smoke <- probsens(matrix(c(215, 1449, 668, 4296),
dimnames = list(c("BC+", "BC-"), c("Smoke+", "Smoke-")), nrow = 2, byrow = TRUE),
type = "exposure", reps = 20000,
seca = list("uniform", c(.7, .95)),
spca = list("uniform", c(.9, .99)))
#> ℹ Calculating observed measures
#> ⠙ Assign probability distributions
#> ✔ Assign probability distributions [14ms]
#>
#> ⠙ Simple bias analysis
#> ✔ Simple bias analysis [41ms]
#>
#> ⠙ Incorporating random error
#> ⠹ Incorporating random error
#> ✔ Incorporating random error [123ms]
#>
plot(smoke, "forest_or")
 set.seed(123)
conf <- probsens_conf(matrix(c(105, 85, 527, 93),
dimnames = list(c("HIV+", "HIV-"), c("Circ+", "Circ-")), nrow = 2, byrow = TRUE),
reps = 20000,
prev_exp = list("triangular", c(.7, .9, .8)),
prev_nexp = list("trapezoidal", c(.03, .04, .05, .06)),
risk = list("triangular", c(.6, .7, .63)),
corr_p = .8)
#> ℹ Calculating observed measures
#> ⠙ Simple bias analysis
#> ! Samplings lead to 1801 instances in which
#> sampled cell counts were zero and discarded.
#> ⠙ Simple bias analysis
#> ✔ Simple bias analysis [129ms]
#>
#> ⠙ Incorporating random error
#> ✔ Incorporating random error [36ms]
#>
plot(conf, "prev_exp")
set.seed(123)
conf <- probsens_conf(matrix(c(105, 85, 527, 93),
dimnames = list(c("HIV+", "HIV-"), c("Circ+", "Circ-")), nrow = 2, byrow = TRUE),
reps = 20000,
prev_exp = list("triangular", c(.7, .9, .8)),
prev_nexp = list("trapezoidal", c(.03, .04, .05, .06)),
risk = list("triangular", c(.6, .7, .63)),
corr_p = .8)
#> ℹ Calculating observed measures
#> ⠙ Simple bias analysis
#> ! Samplings lead to 1801 instances in which
#> sampled cell counts were zero and discarded.
#> ⠙ Simple bias analysis
#> ✔ Simple bias analysis [129ms]
#>
#> ⠙ Incorporating random error
#> ✔ Incorporating random error [36ms]
#>
plot(conf, "prev_exp")
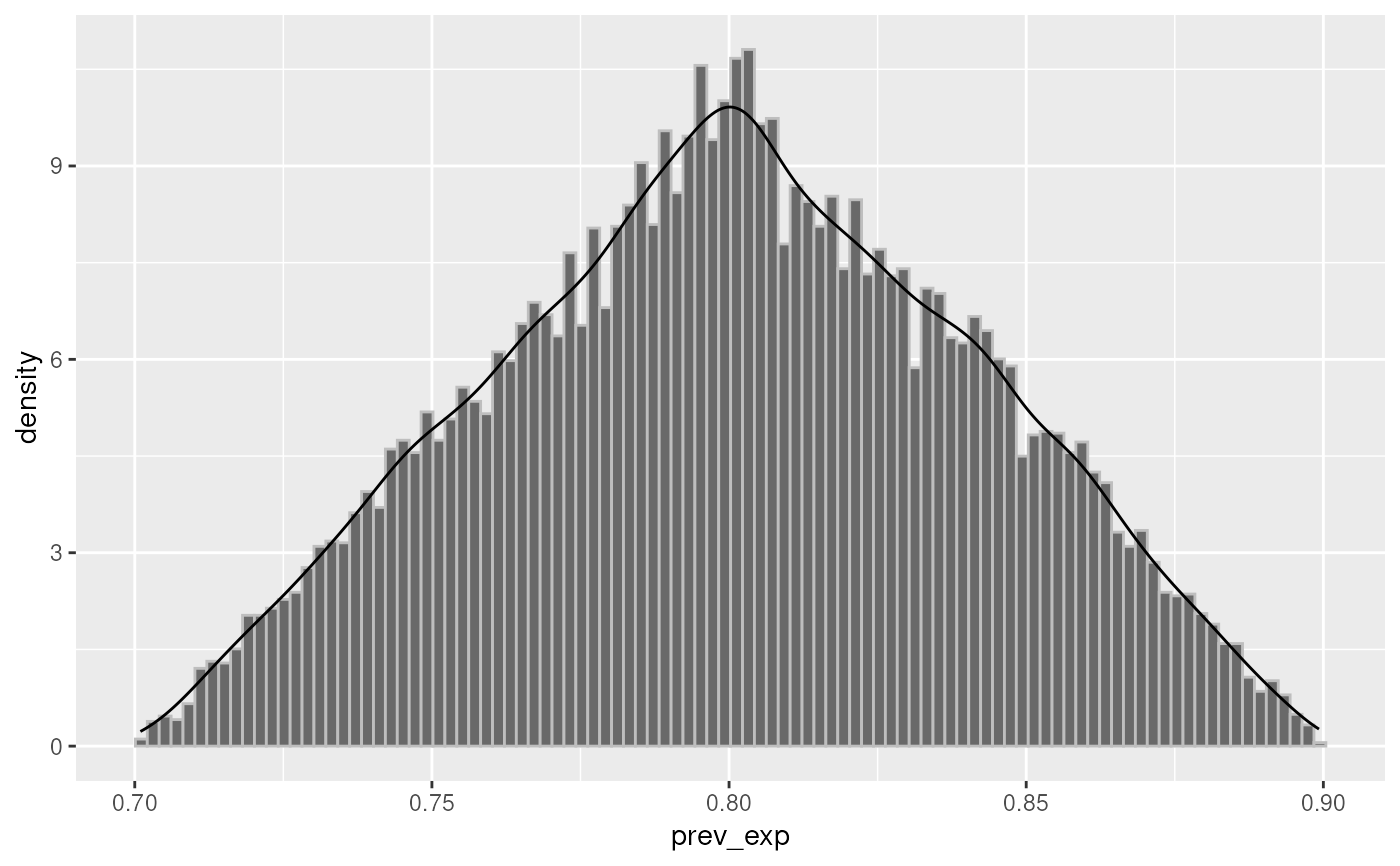 set.seed(123)
inc1 <- probsens.irr(matrix(c(2, 67232, 58, 10539000),
dimnames = list(c("GBS+", "Person-time"), c("HPV+", "HPV-")), ncol = 2),
reps = 20000,
seca = list("trapezoidal", c(.4, .45, .55, .6)),
spca = list("constant", 1))
#> ℹ Calculating observed measures
#> ⠙ Assign probability distributions
#> ✔ Assign probability distributions [10ms]
#>
#> ⠙ Bias analysis
#> ✔ Bias analysis [93ms]
#>
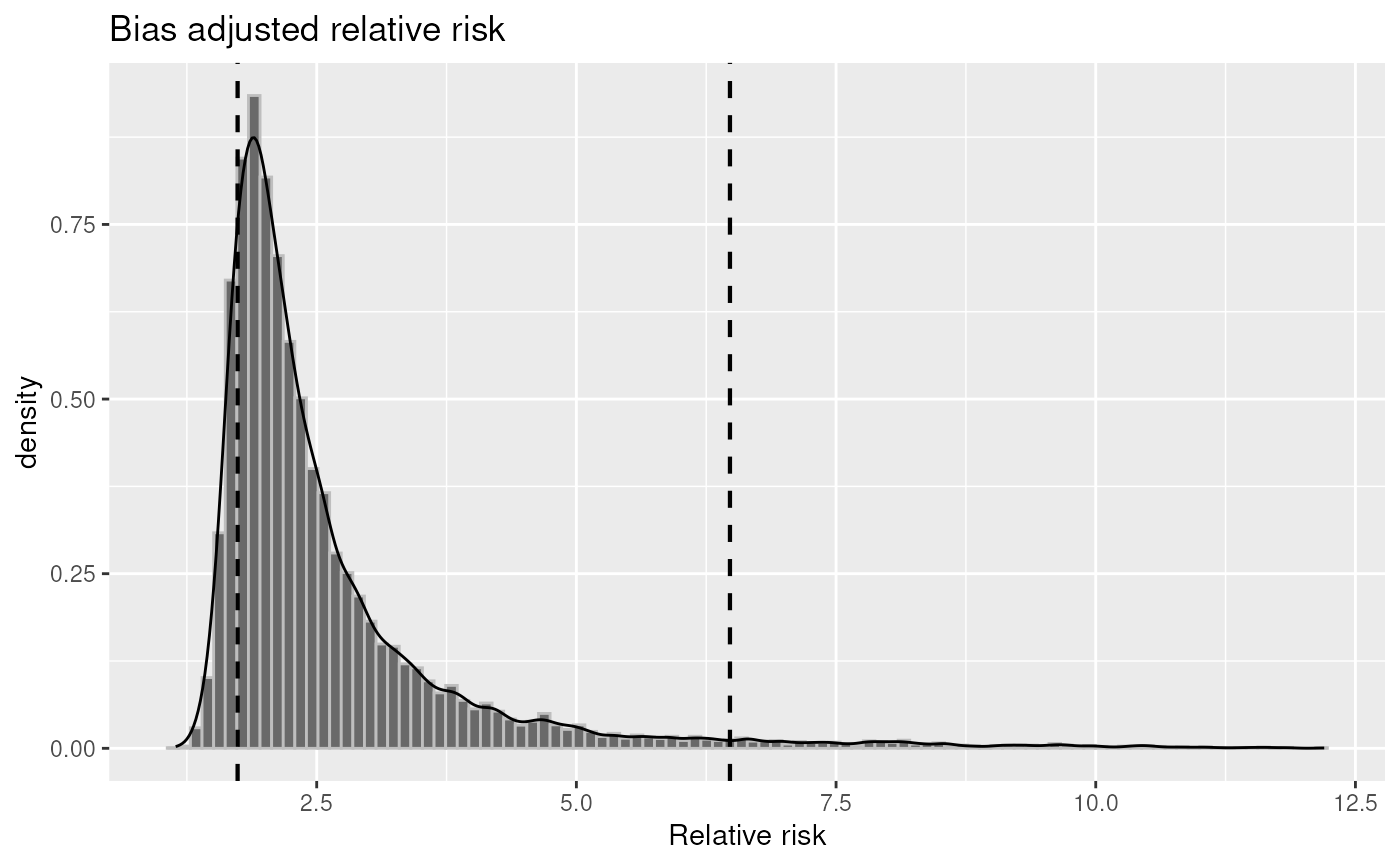
plot(inc1, "irr")
set.seed(123)
inc1 <- probsens.irr(matrix(c(2, 67232, 58, 10539000),
dimnames = list(c("GBS+", "Person-time"), c("HPV+", "HPV-")), ncol = 2),
reps = 20000,
seca = list("trapezoidal", c(.4, .45, .55, .6)),
spca = list("constant", 1))
#> ℹ Calculating observed measures
#> ⠙ Assign probability distributions
#> ✔ Assign probability distributions [10ms]
#>
#> ⠙ Bias analysis
#> ✔ Bias analysis [93ms]
#>
plot(inc1, "irr")
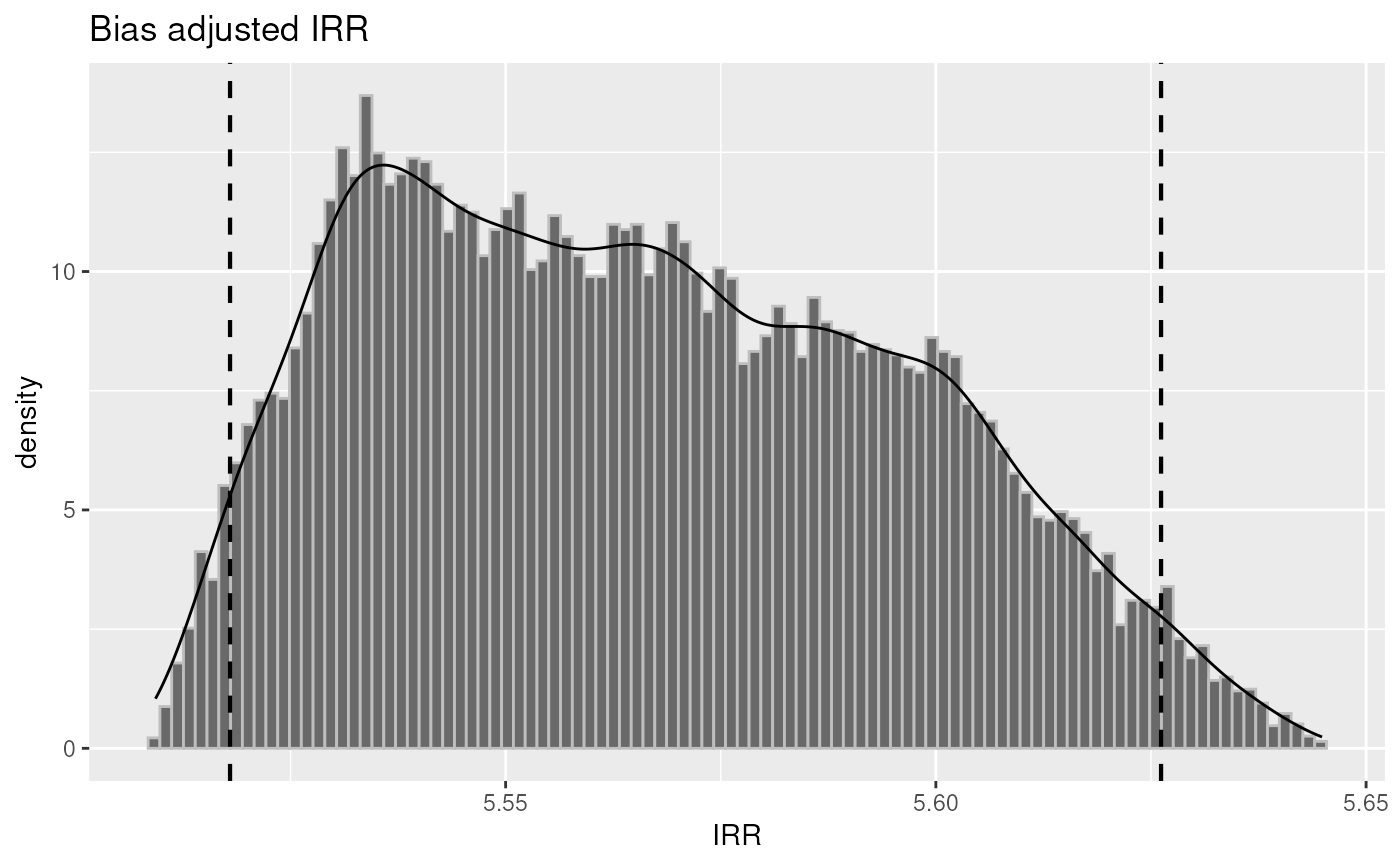 set.seed(123)
inc2 <- probsens.irr.conf(matrix(c(77, 10000, 87, 10000),
dimnames = list(c("D+", "Person-time"), c("E+", "E-")), ncol = 2),
reps = 20000,
prev_exp = list("trapezoidal", c(.01, .2, .3, .51)),
prev_nexp = list("trapezoidal", c(.09, .27, .35, .59)),
risk = list("trapezoidal", c(2, 2.5, 3.5, 4.5)),
corr_p = .8)
#> ℹ Calculating observed measures
#> ⠙ Assign probability distributions
#> ✔ Assign probability distributions [16ms]
#>
#> ⠙ Bias analysis
#> ✔ Bias analysis [116ms]
#>
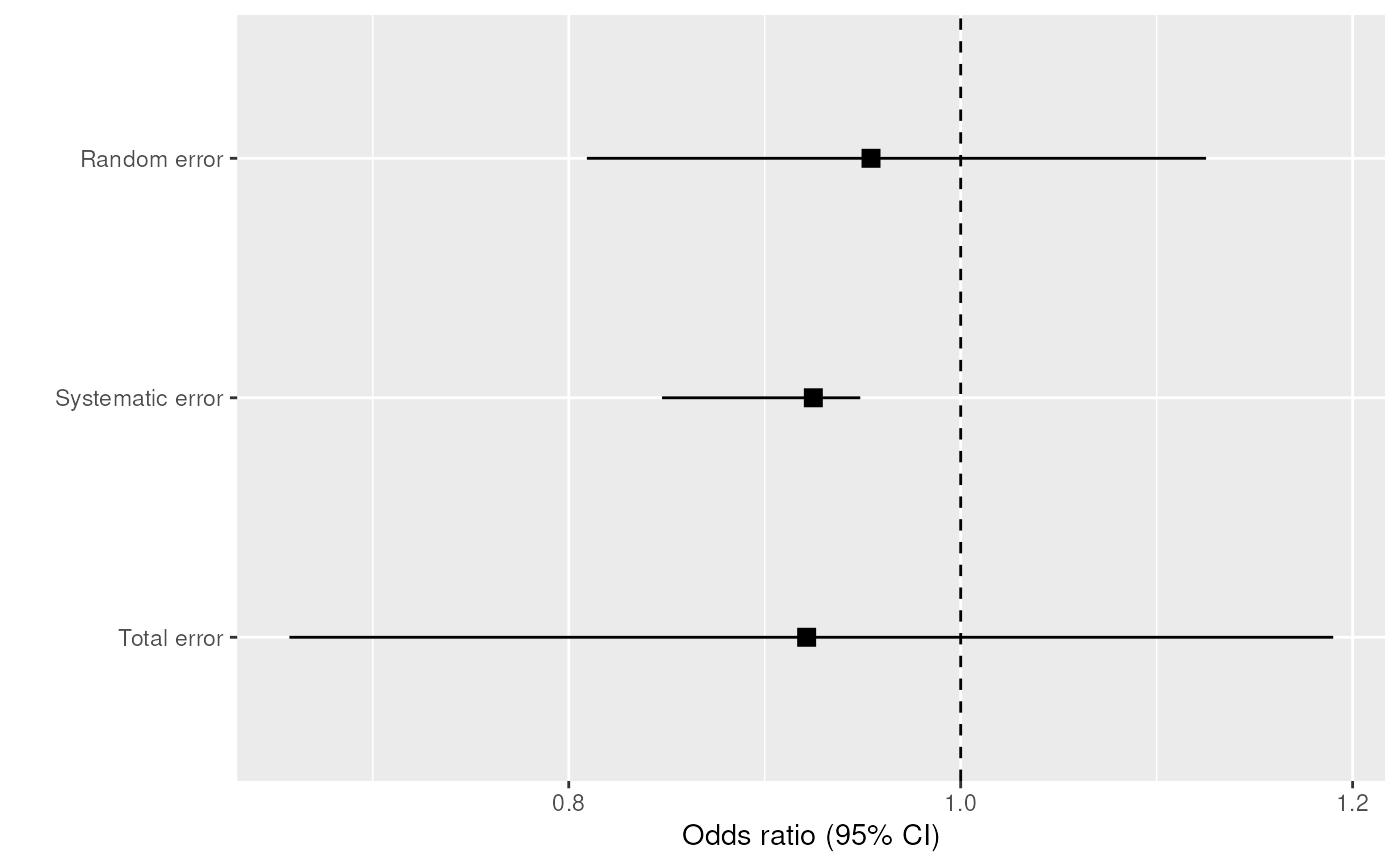
plot(inc2, "risk")
set.seed(123)
inc2 <- probsens.irr.conf(matrix(c(77, 10000, 87, 10000),
dimnames = list(c("D+", "Person-time"), c("E+", "E-")), ncol = 2),
reps = 20000,
prev_exp = list("trapezoidal", c(.01, .2, .3, .51)),
prev_nexp = list("trapezoidal", c(.09, .27, .35, .59)),
risk = list("trapezoidal", c(2, 2.5, 3.5, 4.5)),
corr_p = .8)
#> ℹ Calculating observed measures
#> ⠙ Assign probability distributions
#> ✔ Assign probability distributions [16ms]
#>
#> ⠙ Bias analysis
#> ✔ Bias analysis [116ms]
#>
plot(inc2, "risk")